LIQUITOPY® (LIQUId Tunable microscOPY)
LIQUITOPY® (LIQUId Tunable microscOPY) represents a groundbreaking development of the optical microscope establishing a new paradigm in data collection and image formation with high impact in biophysics. It is designed to attack an open universal question in cellular and molecular biology. Considering the fact that approx. 2m of human DNA are organised in the cell nucleus into chromatin structures that determine the activity and inheritance of our genomes, the key question is the following: what are the local and global 4D (x,y,z,t) chromatin structures in the nucleus that rule the compaction and function of the human genome in the interphase cells and mitotic chromosomes? LIQUITOPY® will allow to elucidate the relationship between chromatin higher-order structure and function at single cell level.
This innovative approach has the merits of 1) the realization of a brand new multiscale microscope allowing simultaneous contrast available in the image formation process; 2) introducing a powerful label free approach to study chromatin organization in situ; 3) allowing, at single living cell level, to define the levels of higher order organization of chromatin in intact and living cells to paint a 4D quantitative vision of those organizational motifs that today are not completely demonstrated keeping our knowledge in midstream between a hierarchical and a brand new vision; 4) the introduction of a new way to form microscopy images within a liquid approach. LIQUITOPY® will pave a new window to study the delicate relationship occurring between structure and function for chromatin-DNA and other macromolecules at the single cell level under living conditions.
During the past 15 years, my group has focused on developing new approaches in advanced optical microscopy to answer key biological questions. LIQUITOPY ® will allow us to tackle a central biophysical question within a long-term vision that beyond its immediate goals bring novel ideas relevant for practical microscopy. Main research lines are related to Nanoscopy, Label Free microscopy and Chromatin-DNA.
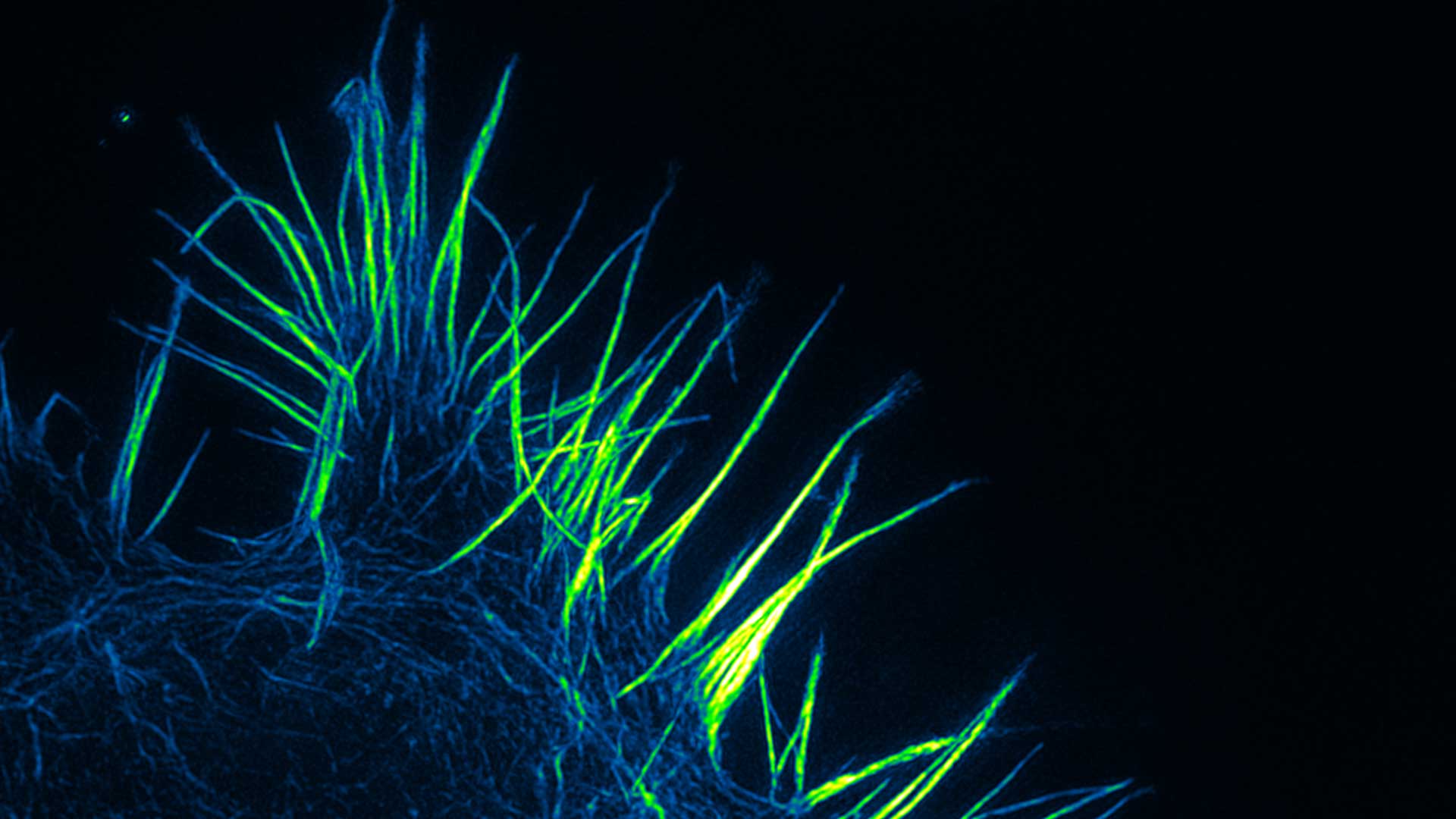
Nanoscopy
The research line related to Nanoscopy deals with the development of novel technologies and instruments for advanced diagnostics at the nanometer scale integrated with focused applications. In the long term, a new paradigm for microscopy development can be envisaged, such as portable multimodal nanoscopes having the potential of being flexible, low power and tunable.
A new generation of microscopes
Such a new generation of microscopes could integrate microscopy, spectroscopy and flow cytometry in the same instrument endowed of all the processing and visualization software on-board.
(Bianchini et al. 2015; Hell et al. 2015; Diaspro & van Zandvoort 2016)
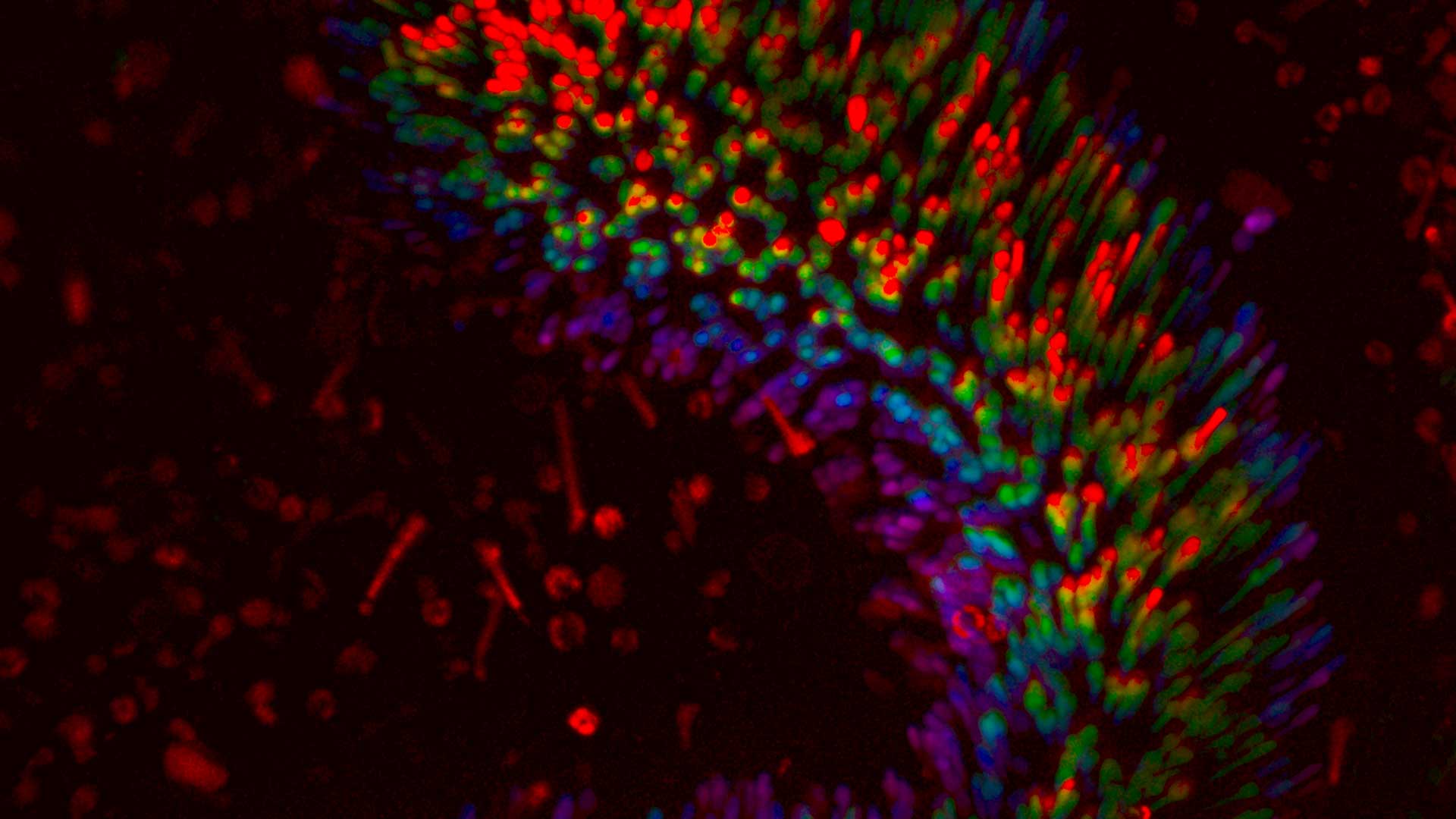
Pump-probe nanoscope
We are developing a pump-probe (or transient absorption) nanoscope. Ultrafast (sub-picosecond) dynamic properties of the sample are investigated with high spatial and temporal resolution, and high sensitivity. Multiphoton and Second Harmonic generation microscopy are integrated to provide a reference within the label free context .
(Bianchini & Diaspro 2008; Teodori et al. 2016)
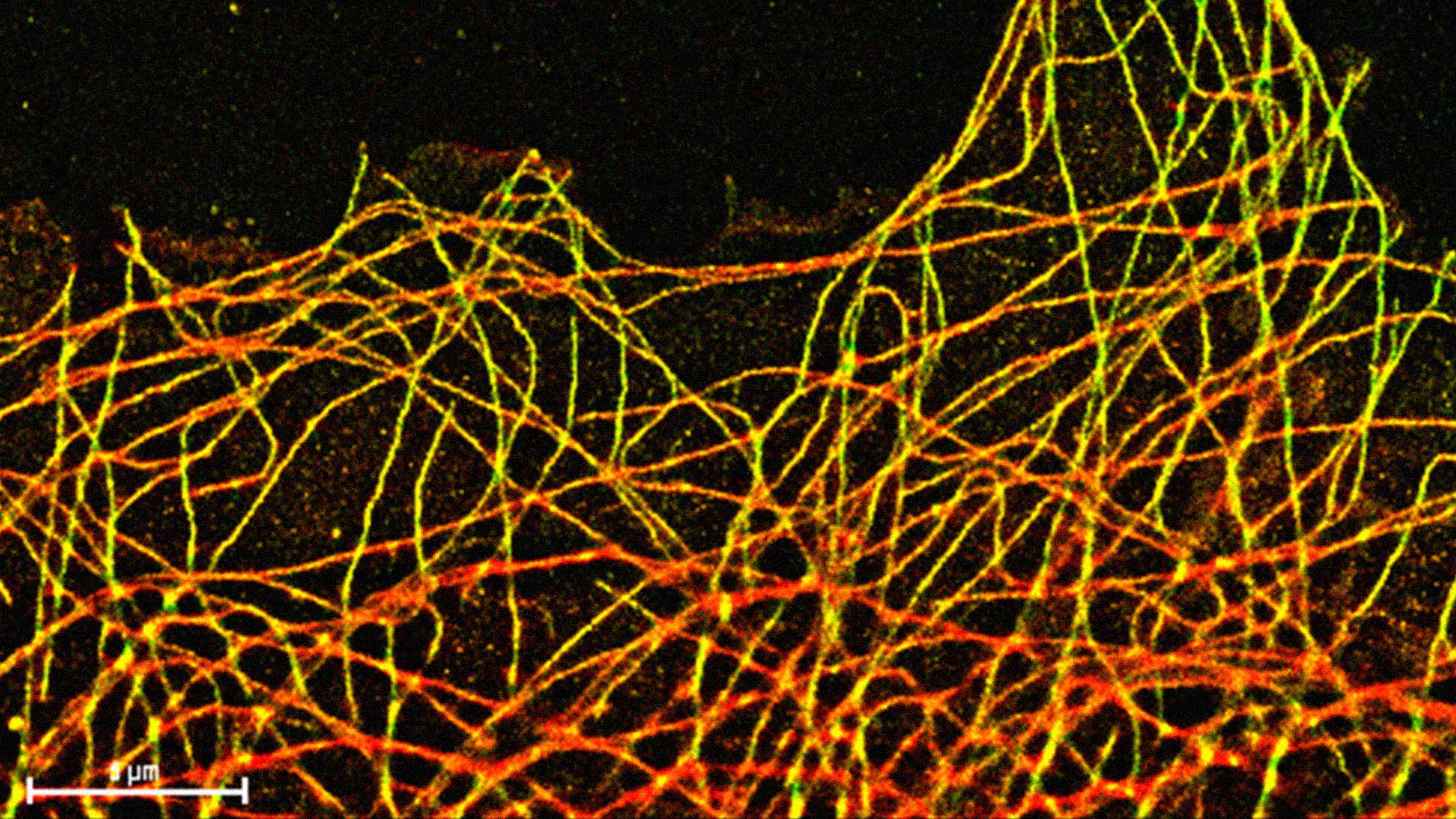
Label free microscopy
In the last decades, non-linear optical processes have captured the attention of life scientists for the development of new super-resolved microscopy techniques(Korobchevskaya et al. 2016). Non-linear optical microscopy goes hand-in-hand with the exploitation of the near-infrared (near-IR) part of the spectrum and was. In order to broaden the range of available targets and provide novel contrast mechanisms in weakly or non-fluorescent samples, absorption-based techniques coming from optical spectroscopy were intensely studied and coupled to with scanning microscopy. This opens the possibility to explore saturation and differential techniques for the circumvention of the diffraction limit also in non-fluorescence-based methods {Liu:2016hq}.
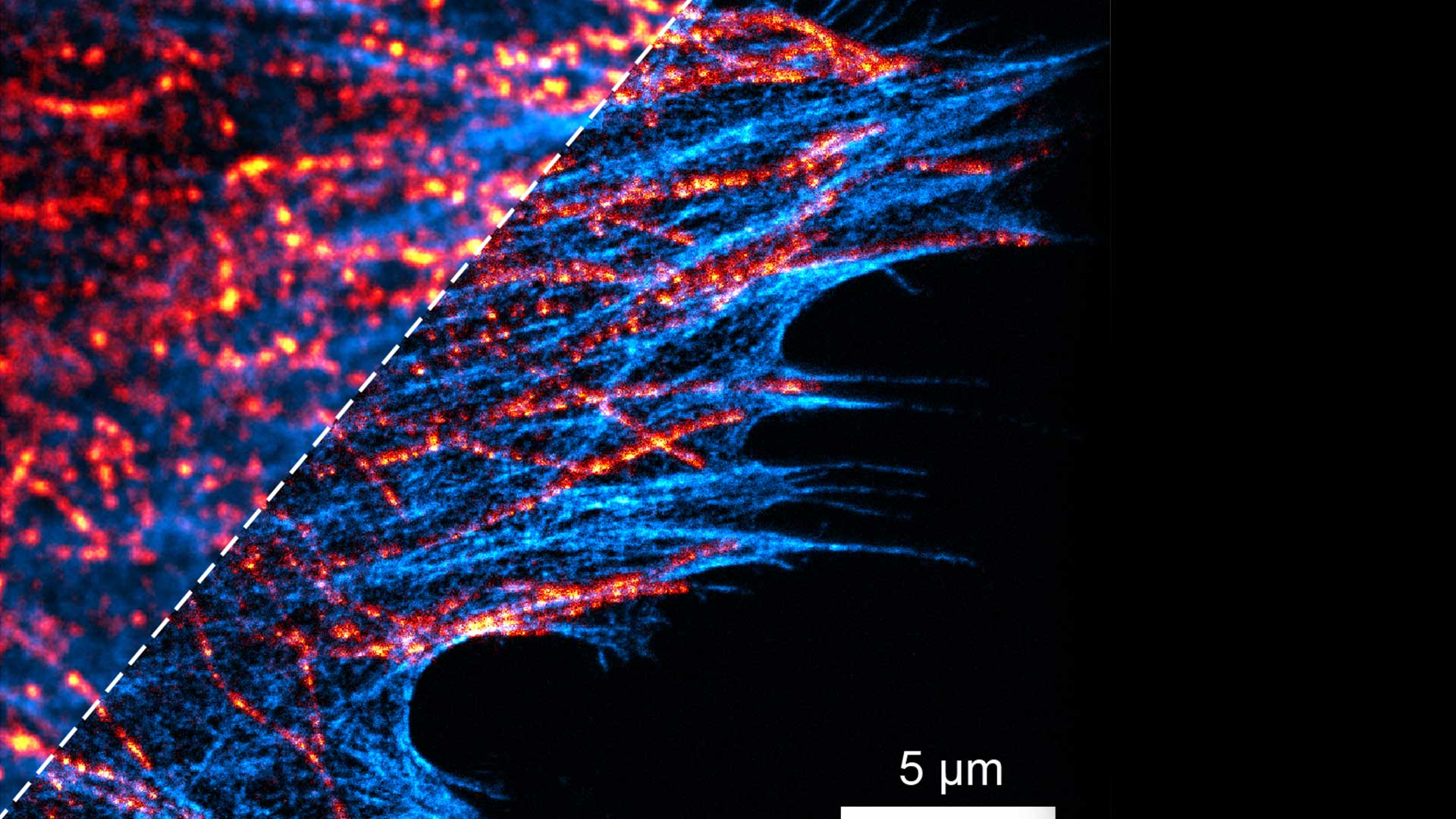
Chromatin - DNA
Considering the fact that approx. 2m of DNA are organised in the cell nucleus into chromatin fibers and clusters that determine the activity and inheritance of our genomes, the key question is the following:
what are the local and global 4D (x,y,z,t) chromatin structures in the nucleus that rule the compaction and function of the human genome in the interphase cells and mitotic chromosomes?
Chromatin relationships
Our studies aim to elucidate the relationship between chromatin higher-order structure and function at single cell level. We have some target pathologies like progeria, achondroplasia and prostate cancer.
(CIT paper Science Ou et al. e quello Nature 4D nucleome)
Publications
For al list of our publications, check the following databases:

Publication list #1
• 01. Scipioni L., Di Bona M., Vicidomini G., Diaspro A., Lanzanò L. (2018) Local raster image correlation spectroscopy generates high-resolution intracellular diffusion maps. Nature Communications Biology. 1:10.
• 02. Vicidomini G., Bianchini P., Diaspro A. (2018) STED super-resolved microscopy. Nature Methods. 15: 173–182.
• 03. G. Sancataldo, L. Scipioni, T. Ravasenga, L. Lanzanò, A. Diaspro, A. Barberis, and M. Duocastella (2017) Three-dimensional multiple-particle tracking with nanometric precision over tunable axial ranges. Optica, 4(3):367–373.
• 04. Duocastella M., Sancataldo G., Saggau P., Ramoino P., Bianchini P., Diaspro A. (2017) Fast inertia-free volumetric light-sheet microscope. ACS Photonics. 4(7): 1797-1804.
• 05. Mazumder N., Qiu J., Kao F., Diaspro A. (2017) Mueller matrix signature in advanced fluorescence microscopy imaging. Journal of Optics. 19(2): 025301
Publication list #2
• 06. Lavagnino Z, Sancataldo G, d’Amora M, Follert P, De Pietri Tonelli D, Diaspro A, Cella Zanacchi F. (2016) 4D (x-y-z-t) imaging of thick biological samples by means of Two-Photon inverted Selective Plane Illumination
Microscopy (2PE-iSPIM). Nature Scientific Reports. 6:23923.
• 07. Castello M., Sheppard C.J.R., Diaspro A., Vicidomini G. (2015) Image scanning microscopy with a quadrant detector. Optics Letters. 40(22): 5355-5358.
• 08. Lanzanò L., Coto Hernandez I., Castello M., Gratton E., Diaspro A., Vicidomini G. (2015) Encoding and decoding spatio-temporal information for super-resolution microscopy. Nature Communications. 6: 6701.
• 09. Viero, G., Lunelli, L., Passerini, A., Bianchini, P., Gilbert, R. J., Bernabò, P., Tebaldi, T., Diaspro, A., Pederzolli, C., Quattrone, A. (2015). Three distinct ribosome assemblies modulated by translation are the building blocks of polysomes. The Journal of Cell Biology 208, 581–596.
• 10. Duocastella M., Vicidomini G., Diaspro A. (2014) Simultaneous multiplane confocal microscopy using acoustic tunable lenses. Optics Express. 22(16): 19293-19301
Publication list #3
• 11. Cella Zanacchi F, Lavagnino Z, Faretta M, Furia L, Diaspro A. (2013) Light-sheet confined super-resolution using two-photon photoactivation. PLoS One. 8 (7), e67667
• 12. Bianchini P., Harke B., Galiani S., Vicidomini G., Diaspro A. (2012) Single wavelength 2PE-STED super-resolution imaging. PNAS – Proceedings National Academy of Sciences USA 109 (17): 6390 – 6393.
• 13. Cella Zanacchi F, Lavagnino Z., Perrone Donnorso M., Del Bue A., Furia L., Faretta M., Diaspro A. (2011) Live-cell 3D superresolution imaging in thick biological samples. Nature Methods. 8 (12): 1047 – 1049.
• 14. Palamidessi A., Frittoli E., Garré M., Faretta M., Mione M., Testa I., Diaspro A., Lanzetti L., Scita G., Di Fiore P.P. (2008). Endocytic trafficking of rac is required for the spatial restriction of signaling in cell migration. Cell. 134(1): 135–147.
• 15. Schneider, M.; Barozzi, S.; Testa, I.; Faretta, M., Diaspro, A. (2005), ‘Two-photon activation and excitation properties of PA-GFP in the 720-920-nm region. Biophysical Journal 89(2), 1346-1352.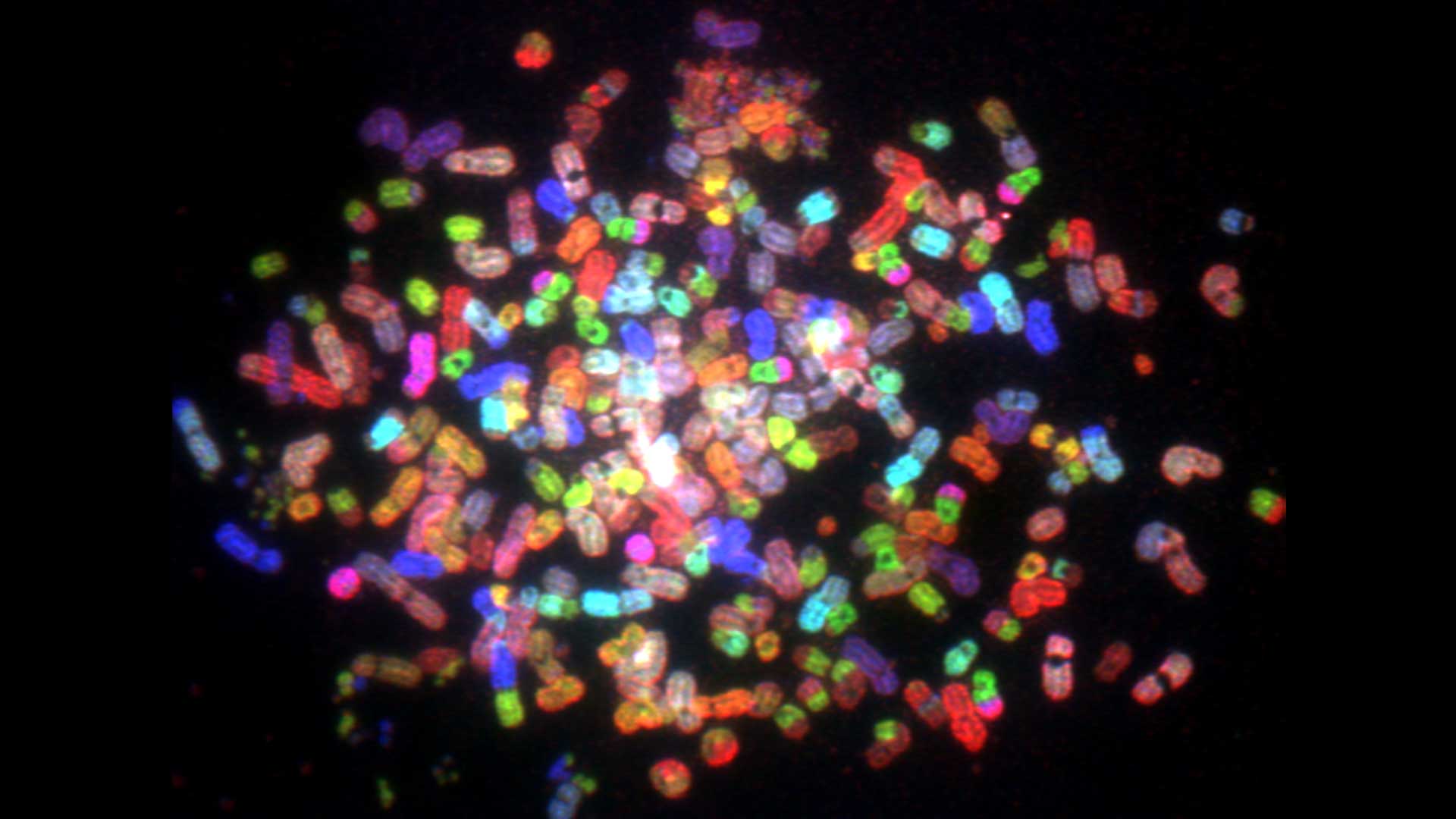
Publication list #4
• 16. Diaspro, A.; Silvano, D.; Krol, S.; Cavalleri, O. & Gliozzi, A. (2002), ‘Single Living Cell Encapsulation in Nano-organized Polyelectrolyte Shells’, Langmuir 18(13), 5047-5050.
• 17. Diaspro, A.; Federici, F., Robello M. (2002), ‘Influence of refractive-index mismatch in high-resolution three-dimensional confocal microscopy.’ Applied Optics 41(4), 685-690.
• 18. Pecere, T.; Gazzola, M. V.; Mucignat, C.; Parolin, C.; Vecchia, F. D.; Cavaggioni, A.; Basso, G.; Diaspro, A.; Salvato, B.; Carli, M., Palù, G. (2000), ‘Aloe-emodin is a new type of anticancer agent with selective activity against neuroectodermal tumors. Cancer Res 60(11), 2800-2804.
• 19. Diaspro, A.; Corosu, M.; Ramoino, P., Robello, M. (1999), ‘Adapting a compact confocal microscope system to a two-photon excitation fluorescence imaging architecture.’ Microscopy Research and Technique 47(3), 196–205.
• 20. Diaspro, A.; Bertolotto, M.; Vergani, L. & Nicolini, C. (1991), ‘Polarized light scattering of nucleosomes and polynucleosomes–in situ and in vitro studies.’, IEEE Trans Biomed Eng July 1991. 38(7): 670-678.
